Figure 1
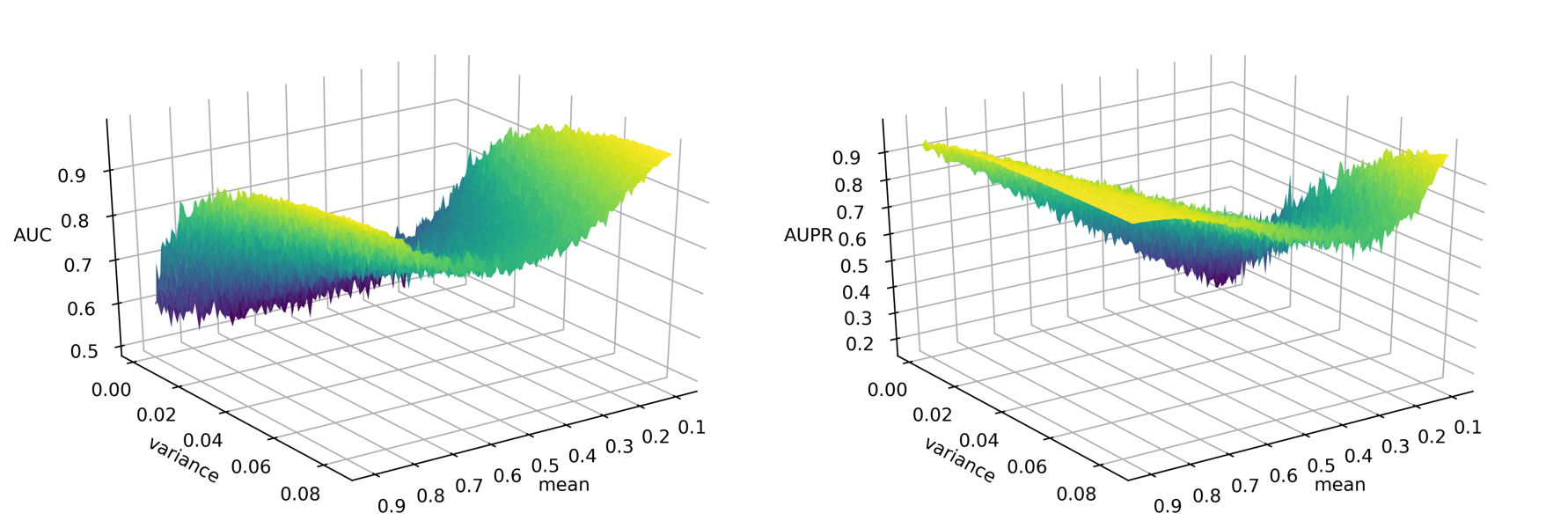
Figure 2
Team 24
Team Members |
Faculty Advisor |
Berk Alpay |
Derek Aguiar Sponsor Pfizer |
sponsored by

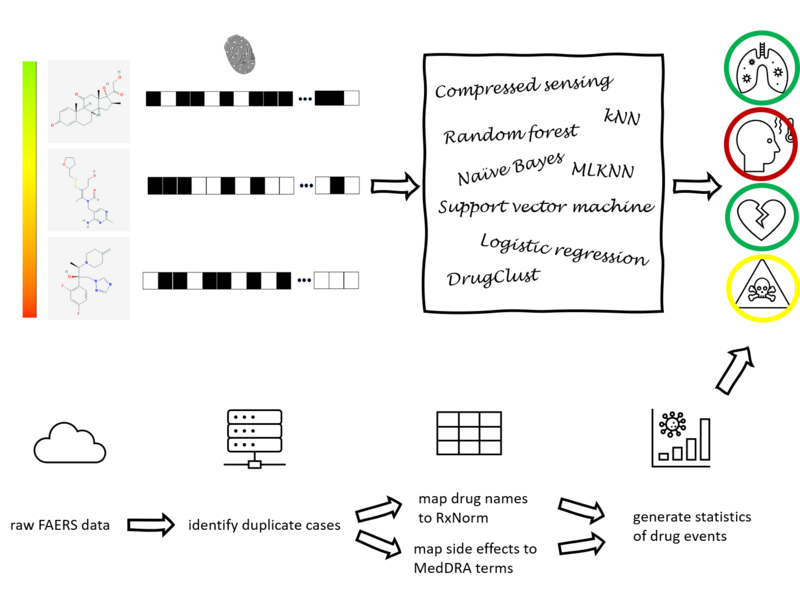
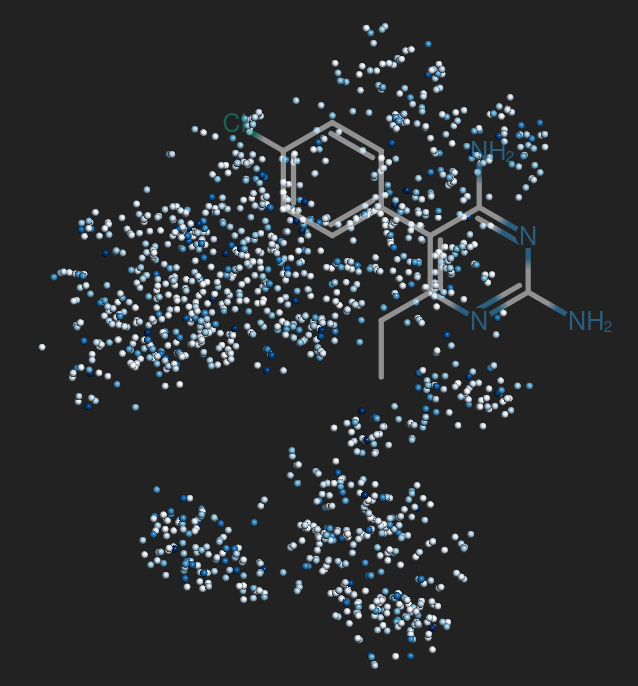
Drug development is an expensive process that requires major investments of time and resources in drugs that may turn out to exhibit severe side effects during clinical trials. To accelerate drug discovery, many machine learning methods have been developed to predict side effects of candidate drugs. However, available data processing pipelines are difficult to execute, evaluations of methods are opaque, and comparisons between them are sparse. In our project, we implemented eight side effect prediction methods and wrapped them into a user-friendly API. To increase the generality of our analyses, we processed two independent datasets. One is the FDA’s FAERS database containing clinical reports of adverse reactions to prescribed drugs. Since these data are input by different doctors, the data is unorganized, inconsistent, and incomplete. We created a pipeline to identify duplicate reports, correct invalid entries using string alignment, and summarize the strength of association between drugs and side effects using a statistic that adjusts for biases in case reporting. We then established a baseline for comparison - a classifier that ignores chemistry and just predicts the empirical frequency of each side effect. Surprisingly, we found that this method is similarly accurate as chemistry-based models as measured by the two most popular metrics in this field. In fact, we developed statistical theory and simulations that explain these results and evaluated our models on other metrics along several different axes. These results led us to the conclusion that a finer-grained approach must be taken to understand the predictions of side effect models. To aid in this, we created a visualization tool that drug developers can use to interpret chemical spaces and uncover trends in model behavior normally hidden by aggregate scores. In summary, our work increases the quality, interpretability, and accessibility of side effect modeling in the pharmaceutical industry.
