Figure 1
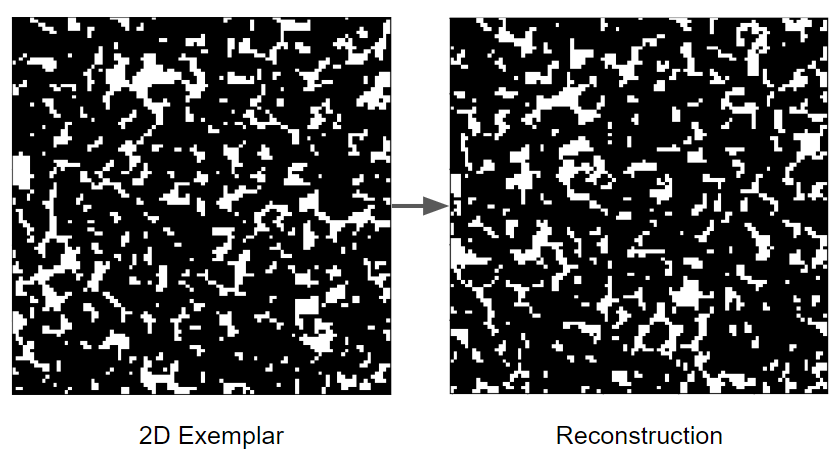
Figure 2
Team 75
Team Members |
Faculty Advisor |
Niccolo Meniconi |
Dr. Xu Sponsor UCONN, Dr. Xu |
sponsored by

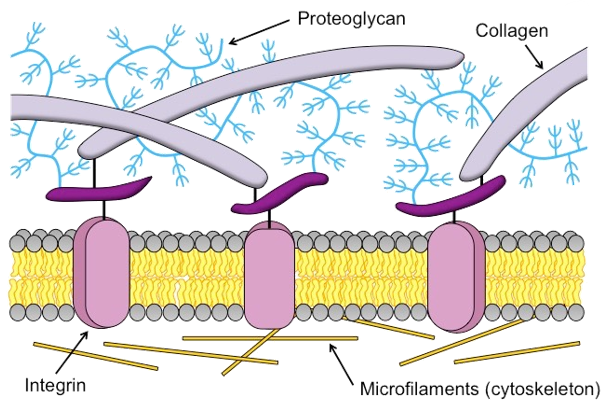
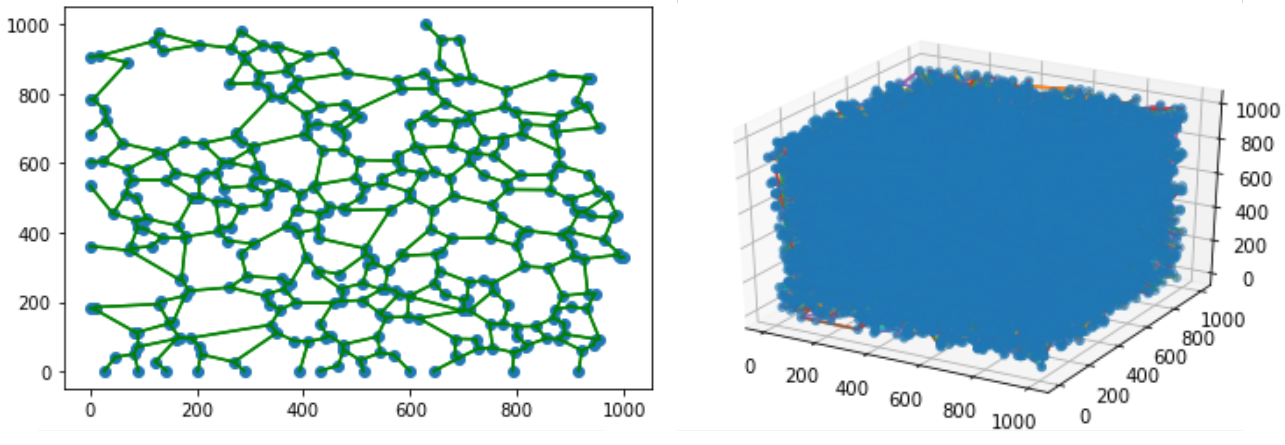
The extracellular matrix (ECM) is a tissue compartment constituted by proteins, glycoproteins, and glycosaminoglycans that assemble to form a complex 3D network. The ECM provides structural and biochemical support to surrounding cells. Despite the important role the ECM plays in forming biological tissues present in many living organisms, there is not a molecular dynamics model which accurately predicts structural behavior in experimental testing. A molecular dynamics model for ECM would provide researchers with a means of predicting ECM behavior with software, rather than with tissue samples. This could decrease the time spent studying diseases of the ECM before a treatment can be developed. Our customers are Dr. Tarakanova and Dr. Xu, who are researchers in the UConn Mechanical Engineering department. We will be developing a computational model of ECM which will allow researchers to simulate ECM dynamics. This project is split into two parts, which we are worked on in tandem. The first part is developing software which, given a 2D image of ECM, can generate a 3D node-edge graph representative of that sample. This node-edge graph will be the input to molecular dynamics software. The second part is developing a computational model of ECM, which, when applied to a 3D node-edge graph generated from an image of ECM, will allow researchers to study the macromechanical properties of that sample.
